Author: Georges Le Goualher
Introduction
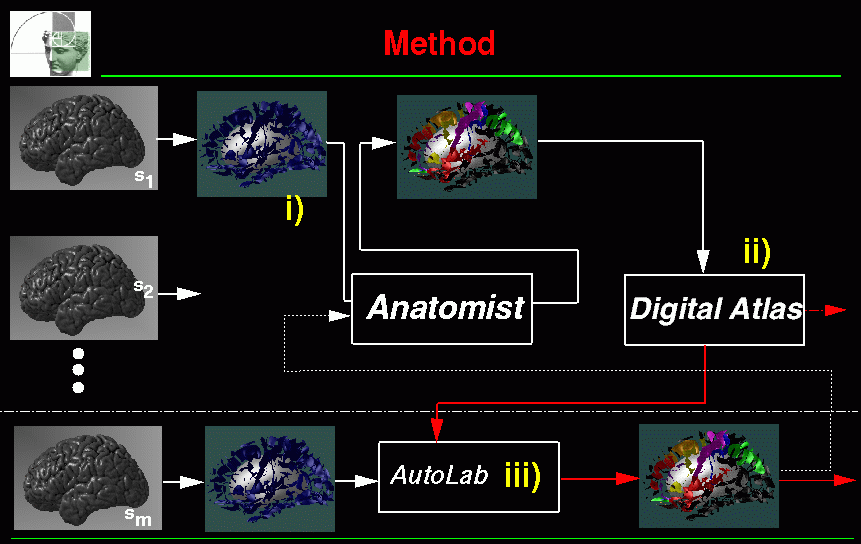
SEAL stands for S ulcus E xtraction and A ssisted L abelling. Figure 1 shows the overview of the application. There are three main parts :
i) Modelling of the cortical sulci (refer to section 2).
ii) Interactive Labelling. An anatomist can interactively label these sulci. This is done using a modified version of Display (refer to section 3).
iii) Automatic Labelling of the main cortical folds. Once a labelling of main cortical folds has been performed on several individuals, Statistical Probability Anatomical Maps (SP_AMs) can be computed. These SP_AMs represent 3D spatial-probability of finding a particular sulcus in the Talairach space (refer to section ). There is one SP_AM per sulcus. Based on these spatial priors an automatic labelling can be performed (refer to section 8).
For data already available and work done within the ICBM project refer to section 6. For related Papers and Reports refer to section 10.
Overview
SEAL script
SEAL is a perl script :
~georges/public_bin/sulcus_graph.pl
command:
sulcus_graph.pl final_t1.mnc surface.obj classification.mnc output_dir
Input Data :
- final_t1.mnc:
3D T1 MRI volume registered and resampled in the MNI-Talairach coordinate system ( c.f http://www.bic.mni.mcgill.ca/~louis/mri_segmentation/ , [2]).
- surface.obj:
Ouput of cortical surface segmentation (i.e. output of ~david/public_bin/cortical_surface.pl, [3] or grey matter surface output of ~georges/public_bin/cortical_surface_double_hull.pl).
- classification.mnc:
Ouput of tissue classication (i.e. output of classify or classify_clean, [5]).
- output_dir:
Directory where ouput of sulcus_graph.pl should be stored.
NOTE : sulcus_graph.pl has been applied to the ICBM database. Output are stored under : /data/icbm/mri1/subjects/00XXX/seg/auto/mni_icbm_00XXX_t1_sulci
For informations on this ICBM database refer to : MNI ICBM MRI DATA BASE
Output Data :
sulcus_graph.pl generates 6 object files and one sulgraph file which are stored under the output_dir directory 1. As an example lets consider ICBM 00100 subject. For this subject, output files are stored under :
/data/icbm/mri1/subjects/00100/seg/auto/mni_icbm_00100_t1_sulci
These ouput files are :
-
G_A_L_L.obj (Graph_Arcs_Left_Line). Contains a set of curves (or ``lines’'). Each curve represents the superficial trace of a particular cortical fold (left hemisphere only).
-
G_A_L_Q.obj (Graph_Arcs_Left_Quadmesh). Contains a set of surfaces (or ``quadmesh’'). Each surface represents the median surface of a cortical fold (left hemisphere only).
-
G_A_R_L.obj ( Graph_Arcs_Right_Line). Equivalent of G_A_L_L.obj for the right hemisphere.
-
G_A_R_Q.obj (Graph_Arcs_Right_Quadmesh). Equivalent of G_A_L_Q.obj for the right hemisphere.
-
G_VERTEX_L.obj (Graph_Vertex_Left). Contains all curve junctions. Each junction is represented by a 3D point (left hemisphere only).
-
G_VERTEX_R.obj (Graph_Vertex_Left). Equivalent of G_VERTEX_L.obj for the right hemisphere.
-
GRAPH_sulci.sulgraph. This is a sulgraph data format file. This file stores all the informations already stored in the previous object files plus other additional data described in the following paragraph.
-
LAB.mnc.gz, POT1.mnc.gz, POT2.mnc.gz and POT3.mnc.gz are minc files containing LABels and POTentials (from which forces are derived). These files are used only for post-modelling 9. In general you don’t need them.
GRAPH_sulci.sulgraph :
The .sulgraph structure is an attributed relational graph structure that I’ve defined for my application 2. A graph contains two main features : Arcs and Vertices 3.
-
Arc : contains a parametrical surface representing the median surface of a fold. Coordinates of 3D points belonging to this surface are expressed in MNI-Talairach world coordinates. We also store :
- length. This is the length of the sulcus superficial trace.
- mean depth. Mean depth of the fold.
- orientation. The fold’s median surface is approximated by a plane (using a least square approximation). The normal vector of this plane is used to represent the orientation of the fold.
- gravity center. The location of the sulcus median surface’s gravity center is computed.
- labels. A set of labels are also assigned to an arc (hemisphere, lobe, name of the fold, sulcus’s type i.e.: main, secondary, tertiary).
- probability vector. We also store a probability vector for each arc. Each component of this probability vector represents the probability that the associated median surface belongs to a given sulcus. These probabilities are computed from SP_AMs already available, i.e. computed off-line (refer tohttp://www.bic.mni.mcgill.ca/~georges/papers/vbc_camread/vbc_camread.html )
-
Vertex : has a 3D location; the vertex contains the list of arcs which connect themselves at this location.
Interactive Labelling
The goal of the Interactive Labelling is to label the sulci of interest. Once this labelling has been performed on several individuals, it’s possible to compute sulcus SP_AMs. The interactive labelling is performed using an modified version of Display called Display.geo (see figure 2). The only difference between Display and Display.geo is that Display.geo contains a specific menu for sulcal labelling 4.
Application
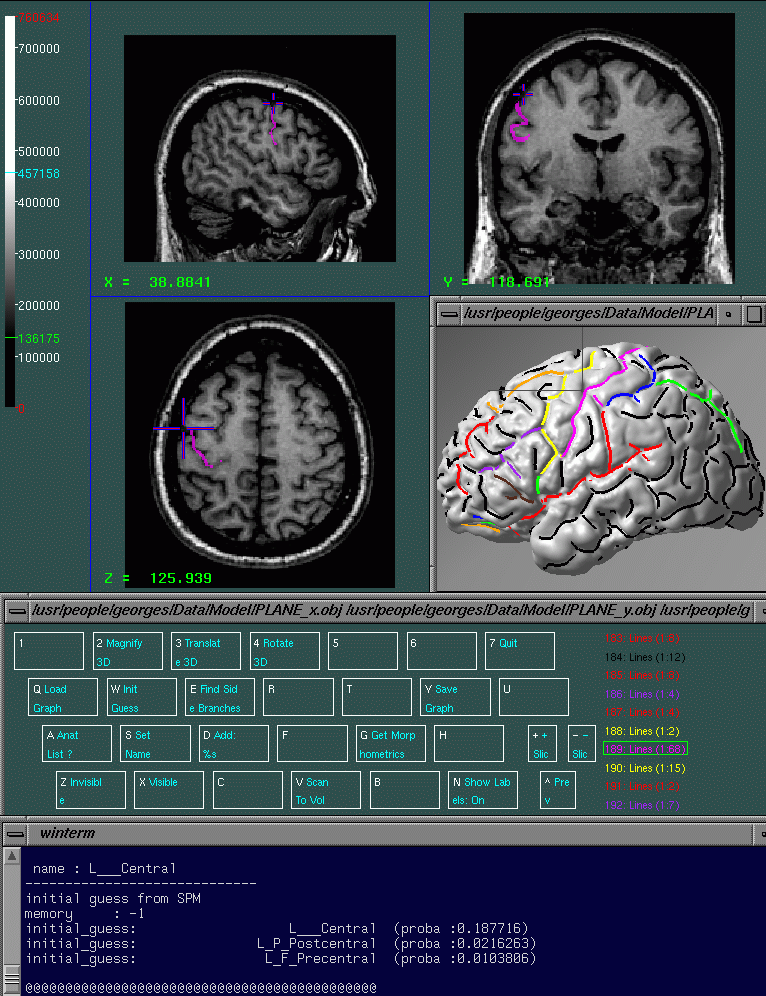
Before Starting: disp.geo alias
You may use directly ~georges/Source/Display/Display.geo for interactive labelling, but I strongly suggest you create a disp.geo alias :
alias disp.geo \verb|~|georges/Source/Display/Display.geo \ ~georges/Data/Model/PLANE_x.obj \ ~georges/Data/Model/PLANE_y.obj \ ~georges/Data/Model/PLANE_z.obj \ ~georges/Data/Model/little_cortex.obj’
Then when you start the command disp.geo you can see that this command start Display.geo and that in this version there is a new button in the main menu : 3D Sulci (associated with the N keyboard key). Several geometrical objects are launched in the 3D window : 3 perpendicular planes and a little cortex model . These objects will be useful to prevent visual overlapping between hemispheres. Of course you can hide an object by :
- selecting it with the mouse
- selecting Objects/Invisible
Running disp.geo
You need :
-
final_t1.mnc. The 3D T1 MRI resampled in the MNI-Talairach space.
-
surface.obj. The cortical surface. Taking advantage of David’s double hull surface extraction I suggest you load the White Matter/Grey Matter Interface where cortical sulci are more visible. Such a White/Grey Interface surface is /data/icbm/mri9/david/White_surfaces/Surfaces/00XXX_81920.obj.gz.
-
G_A_L_L.obj and G_A_R_L.obj
-
GRAPH_sulci.sulgraph
Run :
disp.geo final_t1.mnc surface.obj G_A_L_L.obj G_A_R_L.obj
3D Sulci Menu
Menu
-
Q Load Graph . Load the GRAPH_sulci.sulgraph. This is the first thing to do.
-
W Init Guess . This button performs an initial labeling of all cortical folds based on previously computer SP_AMs (refer tohttp://www.bic.mni.mcgill.ca/~georges/papers/vbc_camread/vbc_camread.html for details). Use the keyboard arrow to select the object number 5 (G_A_L_L.obj). Click on Init Guess : on the window appears :
Continue ? (answer by yes or no)'** enter yes **Enter proba_threshold and depth_threshold:’
enter 0.1 0. All sulci having a probability vector such that its maximum component is superior to 0.1 are labelled. Note that this function works only if G_A_L_L.obj, G_A_R_L.obj, G_A_L_Q.obj or G_A_R_Q.obj are selected. -
E Find Side Branches . Label a non-terminal segment as ``side branche’’ if there is angle close to 90 degrees between this segment and its neighboring sulci. This button is not used in general as its output is not really good.
-
R Compare Two Graphs . Allows to visualy inspect the difference between two graphs.
-
Y Save Graph . Allows to save the graph once the labelling is done.
-
A Anat List? . Display in the terminal window the list of possible labels with their IDs. For corresponding label on the right hemisphere add 100 to the left hemisphere’s label. Example Central Sulcis Label is 15 for the left hemisphere and 115 for the right hemisphere.
-
S Set Name . this button allows to enter the name of the fold (several folds may constitute a sulcus as defined by an anatomist). You don’t need to enter the full name of the sulcus (wich would be too long) but only a number. Refer to table 1 for correspondance between number/name and colour or use the button Anat List? . Note that the probability associate with the label you enter is set to 1.0. (i.e. maximum confidence in the manual labelling). However the initial state of the probability vector is stored in memory and can be retreived if you set the ``unknow’’ label (i.e. 0) to the curve.
-
D : Add . As said before several folds may constitute a sulcus as defined by an anatomist. In order to accelerate such a labeling, use the Add button to set the same name (current name) to several lines. Pick a curve and press the Add button (or enter D on the keyboard). Note that the probability associate with the label you enter is set to 1.0.
-
G Get Morphometrics Informations related to the selected curve are displayed on the window. For example the length and the depth of the selected fold are displayed. The actual name of the curve is also displayed and some probability informations obtained with the SP_AMs. These informations may help you to find the name of the sulcus in which the selected curve belongs.
-
C Smalls Invisible . All sulcus’ portions having a length inferior to the threshold you are asked to enter becomes invisible. To make them visible again use button Visible (X).
-
V Scan To Vol . Once a curve has been selected it is possible to scan the associated median surface onto the MRI volume. This could help you to figure out the name of the selected curve based on its exact location in the MRI. The Show Labels:On/off allows to make the label visible or not on the MRI. The PopMenu/Segmenting/Clear All Labels allows to remove this scanning.
-
B Unknown Invisible . All black curves (i.e all curves labeled with the Unknown label) become invisible.
Available Labels
The available labels are listed in the following table (1).
Anatomical List
| number | sulcus name | color/scanning_color | scanning_label |
|---|---|---|---|
| 0 | unknown_name | BLACK/CYAN | 4 |
| 1 | L_F_Ascending_Ramus | GREEN | 18 |
| 2 | L_F_Horizontal_Ramus | YELLOW | 6 |
| 3 | L_F_Incisura | DARK_BROWN 5 | 14 |
| 4 | L_F_Inferior_Frontal | PURPLE | 7 |
| 5 | L_F_Middle_Frontal | RED | 1 |
| 6 | L_F_Precentral | YELLOW | 6 |
| 7 | L_F_Superior_Frontal | ORANGE | 99 |
| 8 | L_Fo_Intermedius | GREEN | 18 |
| 9 | L_Fo_Lateralis | BLUE | 3 |
| 10 | L_Fo_Medialis | ORANGE | 99 |
| 11 | L_Fo_Olfactory | DARK_BROWN 6 | 14 |
| 12 | L_Fo_Transversus | RED | 1 |
| 13 | L_P_Intraparietal | GREEN | 18 |
| 14 | L_P_Postcentral | BLUE | 3 |
| 15 | L___Central | MAGENTA | 8 |
| 16 | L___Sylvian | RED | 1 |
| 17 | L_T_Superior_Temporal | GREEN | 18 |
| 18 | L_T_Inferior_Temporal | MAGENTA | 8 |
| 19 | L_T_Inferior_Temporal_Ascending_Limb | YELLOW | 6 |
| 20 | L_O_Lateral | RED | 1 |
| 50 | L_F_Subcentral_anterior | DARK_BROWN | 14 |
| 51 | L_F_Frontal_vertical | MAGENTA | 8 |
| 52 | L_F_Precentral_rostral | BLUE | 3 |
| 60 | L_P_Subcentral_posterior | ORANGE | 99 |
| 70 | L_T_Anterior_Collateral | GREEN | 18 |
| 71 | L_T_Posterior_Collateral | MAGENTA | 8 |
| 72 | L_T_Posterior_Ascending_Superior_Temporal | ORANGE | 99 |
| 73 | L_T_Posterior_Horizontal_Superior_Temporal | YELLOW | 6 |
| 99 | L___ERROR | CYAN/BLACK | 40 |
| 100 | L___Side_Branche | WHITE | 41 |
Examples
There are basically two situations : i) you want to sart the labelling of a sulgraph file ; ii) you want to continue or update the labelling of a sulgraph file .
Starting a labelling
You can do the following:
Run :
disp.geo final_t1.mnc surface.obj G_A_L_L.obj G_A_R_L.obj
For the left hemisphere, select G_A_L_L.obj in the object list and apply :
Init Guess .
`Enter proba_threshold and depth_threshold:’
enter 0.1 0
(for right hemisphere do the same thing on object number 6, i.e G_A_R_L.obj).
Now you can see that a subset of lines have been colored (other lines are black). All colored lines are the lines that have been labeled based on spatial priors. To figure out was is the label of a curve select this curve with the mouse, use Get Morphometrics and check in the terminal window its name (ex. name : L___Central). Refer to table 1 for correspondance between number/name and colour.
Now you can start to update the actual labeling :
-
pick a curve
-
click on Get Morphometrics : some informations related to the selected curve are displayed on the terminal window. These informations may help you to figure out the name of the selected curve.
-
Scan To Vol : once a curve is selected it is possible to scan its associated median surface on the MRI volume. This may help you to figure out the name of the selected curve based on its exact location on MRI.
-
Set Name : this button allows to enter the sulcus name of the fold (several folds may constitute a sulcus as defined by an anatomist). You don’t need to enter the full name of the sulcus (wich would be too long) but only a number. Refer to table 1 for correspondance between number/name and colour.
-
Add : If a sulcus is composed of several lines, use the Add button to give the same name (current name) to several lines .
-
Save Graph : this is used to save your labelling. You are asked to enter a sulgraph filename (example: myname_subjectID.sulgraph). Note that you can use the same name as the input sulgraph file is you want to overwrite existing file.
Updating a labelling
In this case you have already a sulgraph file containing a labelling and you want to a) see what has been already done; b) continue the labelling.
Apply Init Guess to the appropriate object (i.e. G_A_L_L.obj, G_A_R_L.obj, G_A_L_Q.obj or G_A_R_Q.obj).
`Enter proba_threshold and depth_threshold:’
enter: 0.9 0
Here the probability threshold is equal to 0.9. Therefore only curves having a probability vector such that one component is superior to 0.9 are labelled. Considering the fact that Set Name and Add buttons set to 1.0 the associated name’s probability , these labels are those which were entered manually (i.e. they correspond to a manual labelling). Then after application of Init Guess with this 0.9 probability threshold, you can see what has been already done. You can then follow the scheme described in the previous paragraph to continue and update this labelling.
Computing New Sulcal Probability Anatomical Map (SP_AMs)
Once the labelling has been performed on several sulgraph files, it’s possible to compute new SP_AMs using :
~georges/public_bin/mm_sulspam
mm_sulspam is used like this 7 :
mm_sulspam [<options>] <file:list_of_sulgraph> \ <inputfile:mni_template><output_basename:SPAM>
As an example 8:
mm_sulspam proto_list_51.txt \ /data/icbm/mri1/models/average_305_mask_1.00.mnc \ /data/icbm/sbd/seal/Spam/Spams_marita_51/L___Central.mnc \ -clobber -sulind 15 -proba 0.9;
was used to generate the SP_AM of the left central sulcus (-sulind 15) which were manually labelled (-proba 0.9) from 51 manually labeled graphs. ``proto_list_51.txt’’ is a text file (the actual file is stored under ~georges/Source/Minctools/Mm_sulspam) which contains the list of input sulgraph files:
proto_list_51.txt /data/icbm/sbd/seal/00100/Sul_graph/lab_marita.sulgraph /data/icbm/sbd/seal/00101/Sul_graph/lab_marita.sulgraph … /data/icbm/sbd/seal/00157/Sul_graph/lab_marita.sulgraph /data/icbm/sbd/seal/00158/Sul_graph/lab_marita.sulgraph
Figure 4 shows an example of such a SP_AM.
SPAMs (Statistical Probability Anatomical Maps )
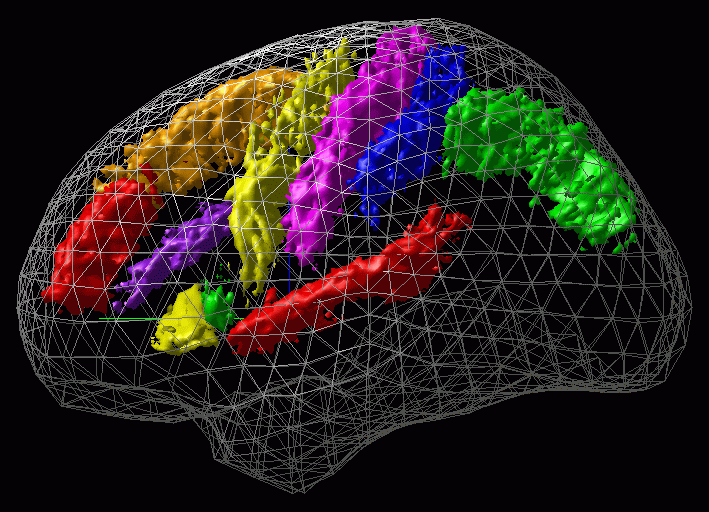
The ICBM Project
The first phase of the ICBM project had consisted in studying spatial variability of anatomical structure in the MNI-Talairach referential frame in order to construct Statistical Probability Anatomical Maps (SP_AMs) of main anatomical structures . This section present the work done within this context giving also practical informations in order to internally retrieve data (the utimate goal of ICBM being to share these data). A complete list of most recent SP_AMs is stored in : ~georges/Source/Minctools/Include/new_tab_of_SPM.text.
Previously Available Data (Caramanos 1)
These data were obtained before SEAL was developped. These data represent spatial variability of sulcal superficial traces while SEAL’s data (refer to 6.3) represent spatial variability of sulcal median surfaces. I strongly suggest to not use these data as new data are now available (refer to 6.3).
A trained neuroanatomist, blinded as to hemispheric location of the sulcus by reflection of hemisphere, identified and manually labelled the superficial trace of a restricted subset of 16 sulci on 51 brains : (a) the Central Sulcus and (b) the Sylvian Fissure; (c) the Superior, (d) the Middle, and (e) the Inferior Frontal Sulci; (f) the Ascending and (g) the Horizontal Rami of the Sylvian Fissure, as well as (h) the Incisura (located between these other two rami); (i) the Olfactory Sulcus as well as (j) the Sulcus Lateralis, (k) the Sulcus Medialis, (l) the Sulcus Intermedius, and (m) the Sulcus Transversus (all found along the orbitofrontal surface of the brain); (n) the Precentral and (o) the Postcentral Sulci; and finally (p) the Intraparietal Sulcus. These data were used to construct associated sulcal SP_AMs, representing the likelihood of finding a particular structure at each 3-D stereotaxique coordinates. These SPAMs are stored under :
sulcus/Tracing/Sulci/Sulcal_Probability_Maps/
You can find the list of these SP_AMs in the file :
~georges/Source/Minctools/Include/tab_of_SPM.text.
The list of subjects that were labeled can be found in table 2 9.
| a | 00100 | 00101 | 00102 | 00103 | 00104 | 00105 | 00106 | 00107 | 00108 | 00109 |
|---|---|---|---|---|---|---|---|---|---|---|
| b | 00110 | 00111 | 00112 | 00113 | 00114 | 00115 | 00116 | 00117 | 00118 | 00119 |
| c | 00120 | 00121 | 00122 | XXX | 00124 | 00125 | 00126 | 00127 | 00128 | XXX |
| d | XXX | 00131 | XXX | 00133 | 00134 | 00135 | XXX | 00137 | 00138 | 00139 |
| e | 00140 | 00141 | 00142 | 00143 | 00144 | 00145 | 00146 | XXX | 00148 | 00149 |
| f | 00150 | 00151 | 00152 | XXX | XXX | 00155 | 00156 | 00157 | 00158 | XXX |
4#2
Emmanuel PROCYK’s Labeling
We have also at our disposal several SP_AMs that were computed from a set of sulgraph files labeled by Emmanuel PROCYK (in collaboration with Michael PETRIDES). These SP_AMs correspond to the frontal lobe sulci : middle frontal s., superior frontal s., inferior frontal s. and also the precentral s., the medial precentral s., the central s., the subcentral anterior sulcus, the subcentral posterior sulcus. 51 brains were studied and the associated sulgraph were labeled. The file /data/icbm/sbd/seal/README contains the list of subjects that were labeled (it is the same list as the one given in table 2). Each labeling is stored in a /data/icbm/sbd/seal/00xxx/Sul_graph/lab_manu_auto.sulgraph.gz file. Computed SP_AMs are no more available because this labelling was updated by Marita SCHMITT.
Marita SCHMITT’s Labeling
The labels of Caramanos and Emmanuel Procyk were then transfered to the lab_marita.sulgraph files and checked by Marita.
This set of 51 sulgraph files contains the most complete sulgraph labeling. These files are under
/data/icbm/sbd/seal/00XXX/Sul_graph/lab_marita.sulgraph . Associated SP_AMs are stored under /data/icbm/sbd/seal/Spam/Spams_marita_51. These SP_AMs represent the data that should be available within the ICBM project. A complete list of most recent SP_AMs is stored in :
~georges/Source/Minctools/Include/new_tab_of_SPM.text.
These SP_AMs represent the 3D distribution of the sulcus median surface (not just the superficial trace distribution as done by Caramanos).
Questions
Where are the SP_AMs?
The file ~georges/Source/Minctools/Include/new_tab_of_SPM.tex contains the filename and path of the SP_AMs.
Most of the SP_AMs are under : /data/icbm/sbd/seal/Spam/Spams_marita_51
Some of them are also under :
/data/npsych/home/sulcus/Tracing/Sulci/Sulcal_Probability_Maps
What should I do if the label I want to use is not available?
Unfortunatly this is not straightforward as there are several modifications to make in the code. Therefore the official ``maintainer’’ will have to update the code and recompile the library LibSul, Display.geo and then mm_suldata.
- in enumeration.h (LibSul/enumeration.h) change the appropriate L___NotSetYet_XX with your new name. Do the same for the right hemisphere i.e. change the corresponding R___NotSetYet_XX with your name.
- in extern char GLOBAL_tab_of_name[NUMBER_SPM_VOL][SIZ_SUL_NAME] (LibSul/ARCDATA.cc) change the associated line with your name (do it for both hemisphere). GLOBAL_tab_of_name and enum seg_name should HAVE THE SAME SYMBOLS.
- in label_graph.cc (LibSul): change all occurence of the L/R___NotSetYet_XX with L/R_?_YOURNAME.
- in the same file (i.e. label_graph.cc), in Colour Global_tab_of_color_name(seg_name name) choose the color you want to associate to your sulcus :
case R___NotSetYet_1 : color = YOURCOLOR; break;
In the case your want to use a new colour (which was not previously used, i.e. which does not appear in table 1), in:
int compute_label_from_color(Colour col, Colour *scanning_col)
put this lines :
if(col == YOURCOLOR){ label = YOURLABEL; *scanning_col = YOURSCANNINGCOLOR; found = YES; //DIFFER }
-
Update /Source/Display/callbacks/georges.c at the section ``Display the anatomical list’', in order to keep the Anat List? button up to date.
-
Update ~georges/Source/Minctools/Include/ANATOMICAL_LIST.txt file
-
Update this file or in its initial Latex version table 1.
Note that the three last actions have no effect on the code, but you need to make them in order to keep everything updated.
Once this is done, recompile the library LibSul, the program mm_suldata (make install) and Display.geo. To recompile Display.geo, you need to do the following (in order to have: setenv SRC_DIRECTORY /data/nil/david/Release) :
cd ~georges/Source/Display source ~david/Source/Makefile_multi_architecture/cshrc.csh; source ~david/Release/cshrc.csh; make Display.geo;
What should I do if a new SP_AM becomes available?
Again it’s the ``maintainer’’ who should do this. If a new SP_AM becomes available you may want to incorporate this information in the computation of new probability vector (i.e. in the computation of new sulgraph file). To do that you have to update ~georges/Source/Minctools/Include/new_tab_of_SPM.text. Example : if the SP_AM of L_T_Superior_Temporal (17) becomes available edit line 17 of ~georges/Source/Minctools/Include/new_tab_of_SPM.text (i.e. add the filename there). Edit also line 117 with the corresponding path for R_T_Superior_Temporal (117). Recompile and install mm_suldata
(i.e. cd ~georges/Source/Minctools/Mm_suldata; make install) . The new version of sulcus_graph.pl will incorporate these new SP_AMs in the computation of the probability vectors.
Automatic Labelling
As said before, automatic labelling of the main cortical sulci is done using prior knowledge for the expected 3D spatial distribution of these entities. Probabilistic identification of a new graph is achieved by integrating each sulcal spatial distribution map over a sulcus’ median surface and assigning it the sulcus name corresponding to the maximum integrated probability value. Quantitative evaluation of the automatic labelling accuracy with regards to the different types of errors (under-estimation of sulcal extend, confusion between adjacent sulci) were computed. Results show that correct identification of eight main cortical sulci studied can be reached in 34 % to 70 % of the cases depending on the level of accuracy needed. For more information on this problem, refer to ~georges/Docs/Postdoc/Article/article.tex and ~georges/Docs/Postdoc/Article/Script.
Post Segmentation
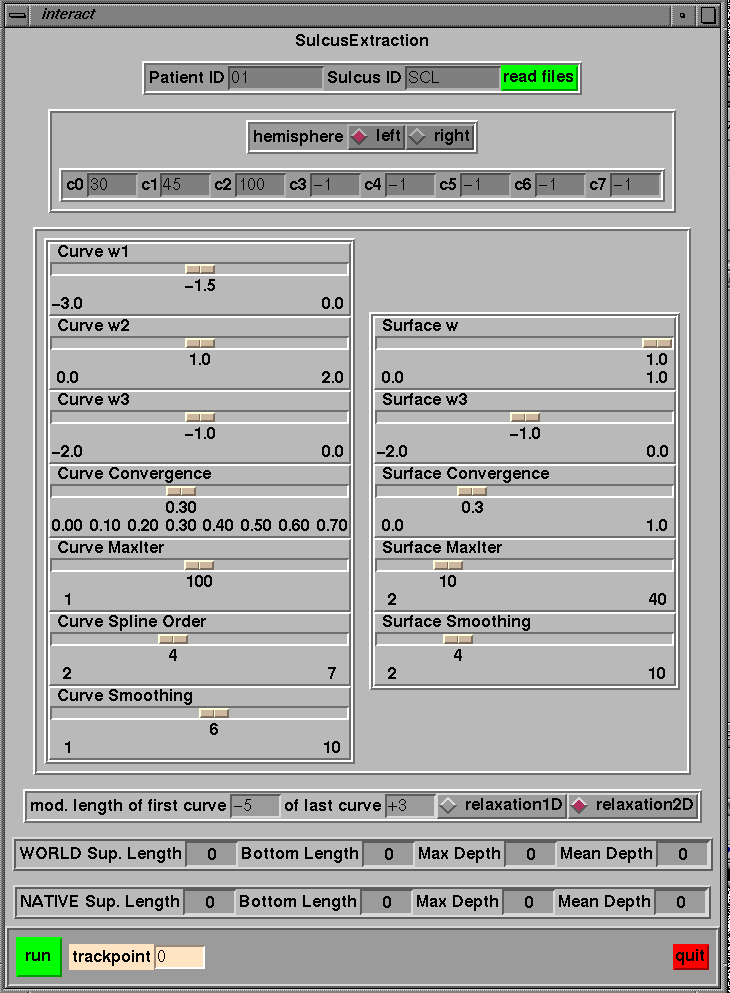
Post-segmentation can be done using ~georges/Source/Minctools/Mm_depth_interac/Tcl/interact. The reason for doing a post-segmentation is that the structural decomposition of the sulcal topography as obtained by SEAL is based on the observation of the external part of the brain. Then a sulcus is generally decomposed in several segments (several portions). Then if it’s needed to model a sulcus as a single element, one need to link the different portions of the sulcus of interest. The following illustration tells the post-segmentation to connect curve number 30, 45 and 100 (of the left hemisphere) to create a simple surface. Moreover the user wants to cut 5 mm at the beginning of the curve and want to add 3 mm at the end of the fold. The user can also change the parameters of the active ribbon. Note that this program need to know where data are stores (i.e. where to find POT1, POT2 etc…), this is defined in interact.h.
Post Segmentation
Georges Le Goualher
1999-12-21